The Microbiome and Human Microbiome Project
Understanding microbial communities and their role in human health through scientific research.
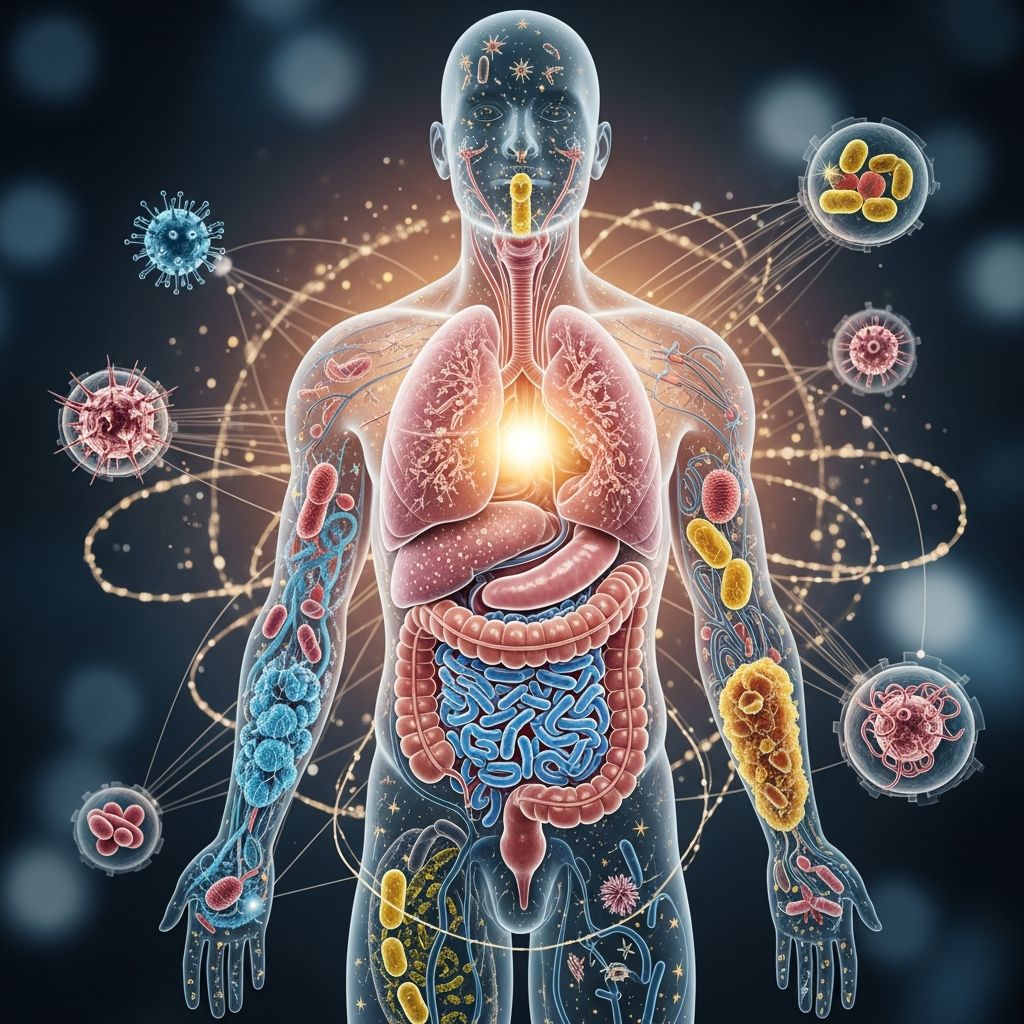
Understanding the Microbiome and the Human Microbiome Project
The human microbiome represents one of the most significant discoveries in modern biology, fundamentally changing our understanding of human health and disease. The microbiome comprises trillions of microorganisms, including bacteria, viruses, fungi, and other microbes that inhabit various sites in the human body, with the gut microbiome being the most extensively studied. These microbial communities play crucial roles in digestion, immune function, metabolism, and mental health. The Human Microbiome Project (HMP), launched in 2008, marked a pivotal moment in microbiome research by establishing comprehensive frameworks and methodologies to characterize these complex microbial ecosystems.
The Human Microbiome Project: Foundations and Objectives
The Human Microbiome Project emerged from a critical need to understand the intricate relationships between human hosts and their resident microbial communities. Before 2008, scientists faced significant limitations in characterizing microbial populations because many microorganisms could not be cultured in laboratory conditions. The HMP revolutionized this landscape through the adoption of culture-independent methods that enabled researchers to identify and analyze microbial communities directly from environmental samples. This technological advancement allowed scientists to bypass the traditional requirement of culturing microorganisms, dramatically expanding the scope and accuracy of microbiome research.
The primary objectives of the Human Microbiome Project included mapping the composition of microbial communities across different body sites, understanding the genetic potential of these communities, and establishing baseline data for what constitutes a healthy microbiome. Researchers collected samples from multiple body sites including the gastrointestinal tract, oral cavity, skin, and urogenital tract, creating comprehensive databases that serve as references for understanding normal microbiome composition and variation among healthy individuals.
Culture-Independent Methods and Technological Advancement
The shift from culture-dependent to culture-independent methodologies represents a fundamental transformation in microbiome research. Traditional microbiology relied heavily on culturing microorganisms on nutrient-rich media, a process that successfully cultivates only a fraction of the microbial world. Estimates suggest that 99% of environmental microorganisms cannot be cultured using conventional laboratory techniques. Culture-independent methods, particularly DNA sequencing technologies and molecular analysis, allowed researchers to detect and identify organisms directly from samples without requiring cultivation.
These advanced techniques include 16S rRNA gene sequencing, which targets conserved bacterial genes to identify bacterial taxa, and metagenomic sequencing, which sequences all DNA present in a sample. These approaches provide comprehensive inventories of microbial communities, revealing the incredible diversity within the human microbiome and enabling researchers to correlate specific microbial compositions with health states and disease conditions.
Probiotics, the Microbiome, and Host Immune Response
Probiotics represent one of the most studied complementary health approaches related to microbiome manipulation. Probiotics are live microorganisms that, when administered in adequate amounts, confer health benefits to the host. The relationship between probiotics, the microbiome, and immune function creates a complex network of interactions that researchers continue to elucidate. When probiotics are ingested, they interact with the existing microbiome and can potentially modulate immune responses through multiple mechanisms.
The host immune response to probiotics involves both innate and adaptive immune pathways. Beneficial bacteria interact with intestinal epithelial cells and immune cells, triggering responses that enhance barrier function, promote beneficial antibody production, and regulate inflammatory responses. The specific mechanisms depend on the bacterial strain, the individual’s existing microbiome composition, and various host factors including genetics, diet, and health status.
Efficacy and Safety of Probiotics: Unanswered Questions
Despite decades of research and widespread commercial availability of probiotic products, significant questions remain regarding their efficacy and safety. The probiotic field faces several challenges that complicate interpretation of research findings and clinical recommendations. One fundamental issue involves standardization—probiotic products vary dramatically in composition, viability, and potency. A probiotic product labeled as containing a specific bacterial species in a certain quantity may deliver vastly different amounts of viable organisms depending on manufacturing, storage, and handling conditions.
Research outcomes also vary considerably depending on the specific bacterial strains tested, dosages used, duration of supplementation, and the health conditions being studied. Some probiotics demonstrate clear benefits for specific conditions like antibiotic-associated diarrhea or certain gastrointestinal disorders, while evidence for many other claimed health benefits remains limited or inconsistent. Rigorous, well-designed clinical trials specifically comparing probiotic interventions to placebo controls are needed to establish definitive efficacy for most applications. Additionally, while probiotics are generally considered safe for healthy individuals, concerns exist regarding their use in immunocompromised populations, and more comprehensive safety surveillance data would strengthen our understanding of potential adverse effects.
Omics Technologies for Understanding Microbiome Function
Modern microbiome research increasingly relies on omics technologies—integrated analytical approaches that examine entire biological systems simultaneously. These technologies include genomics (studying all genes present), metagenomics (analyzing genetic material from environmental samples), transcriptomics (measuring gene expression), proteomics (studying proteins), and metabolomics (examining metabolic products). Omics technologies provide unprecedented insights into how microbiomes function and how probiotics influence microbial communities and host physiology.
Metagenomic approaches allow researchers to assess not only which microorganisms are present but also their genetic potential—what functions they could theoretically perform. Transcriptomic studies reveal which genes microbes actually express under specific conditions, providing information about their functional activity. Metabolomic analyses identify the chemical products of microbial metabolism, linking microbial activity to specific host health outcomes. These integrative approaches help explain probiotic mechanisms of action and predict which individuals might benefit from specific probiotic interventions based on their baseline microbiome composition and metabolic profiles.
Manipulation of the Microbiome by Probiotics
When probiotics are administered, they enter an already-established microbial ecosystem and must compete for resources and ecological niches. The extent to which probiotics successfully colonize and persist in the microbiome varies considerably. Some probiotic strains transiently increase in abundance after ingestion but disappear once supplementation ceases, while others may establish more persistent populations, particularly if they find compatible niches in the microbial community.
Probiotics can influence the microbiome through several mechanisms. They may directly compete with pathogenic organisms for nutrients or adhesion sites. They produce antimicrobial compounds such as bacteriocins that inhibit competing microbes. Additionally, probiotics can alter the local chemical environment, changing pH or oxygen levels in ways that favor beneficial microbes while inhibiting harmful ones. Beyond direct antagonism, probiotics can promote the growth of other beneficial microorganisms through metabolic cross-feeding, where one organism produces compounds that another organism consumes, creating cooperative networks within the microbial community.
Fecal Microbiota Transplantation: A Novel Probiotic Approach
Fecal microbiota transplantation (FMT) represents a unique and increasingly recognized approach to microbiome manipulation. Rather than introducing single or multiple specific bacterial strains, FMT involves transferring entire microbial communities from healthy donors to recipients. This approach proved remarkably effective for treating recurrent Clostridioides difficile infections, achieving cure rates exceeding 90% in many studies, compared to approximately 25% with antibiotic therapy alone.
The success of FMT in C. difficile infections demonstrated that restoring a complex, diverse microbiome could achieve clinical outcomes that targeted interventions could not. This discovery sparked interest in applying FMT to other conditions, though results have been more variable for inflammatory bowel disease, irritable bowel syndrome, and metabolic disorders. The mechanisms underlying FMT efficacy likely involve rapid restoration of microbial diversity and the competitive exclusion of pathogenic organisms by the transplanted community. However, FMT also raises unique considerations regarding donor screening, standardization of procedures, and optimal delivery methods.
Research on Cohousing: Effects on Phenotype and the Microbiome
An intriguing area of microbiome research involves studying how living environments and cohabitation influence microbiome composition and health outcomes. Cohousing research examines whether individuals living in close proximity experience microbiome sharing and whether this affects their health phenotypes. Early studies suggest that cohabitants share greater microbiome similarity than unrelated individuals, and shared environmental factors influence microbial composition more than individual genetics alone in some contexts.
This research has important implications for understanding disease transmission, microbiome resilience, and the role of environmental factors in shaping microbial communities. Studies of families living in the same household, college dormitory residents, and other cohabiting groups provide natural experiments for understanding how physical proximity, shared food sources, and exposure to environmental microbes shape individual microbiomes and potentially influence health outcomes.
Immunomodulatory Properties of Probiotics
A central research focus involves understanding how probiotics exert immunomodulatory effects—how they influence immune system function. Probiotics can enhance innate immune responses by stimulating natural killer cell activity and promoting the production of antimicrobial peptides. They also influence adaptive immunity by promoting the development of regulatory T cells that suppress excessive inflammatory responses, while simultaneously enhancing the development of protective immune responses to pathogens.
Different probiotic strains exhibit distinct immunological profiles. Some strains preferentially promote anti-inflammatory responses, while others enhance pro-inflammatory immunity. This heterogeneity explains why not all probiotics perform identically and why individual strains should be evaluated separately. Understanding these strain-specific immunological effects could enable personalized probiotic selection based on individual health needs and existing immune status.
Current Research Priorities and Knowledge Gaps
The National Center for Complementary and Integrative Health (NCCIH) recognizes microbiome research as a priority area aligned with its strategic mission to advance evidence-based complementary and integrative health approaches. Key research priorities include filling fundamental knowledge gaps about microbiome mechanisms, conducting rigorous clinical trials to establish probiotic efficacy for specific conditions, and developing better methods for standardizing and characterizing probiotic products.
Additional research needs include understanding individual variation in probiotic response, identifying biomarkers that predict who will benefit from specific interventions, and evaluating long-term safety and efficacy of probiotic supplementation. The field would benefit from increased interdisciplinary collaboration between microbiologists, immunologists, gastroenterologists, and other specialists to develop comprehensive models of microbiome-host interactions.
Implications for Whole Person Health
Microbiome research exemplifies the whole person health approach that NCCIH emphasizes in its strategic planning. The microbiome connects biological systems including digestion, immunity, and metabolism with behavioral factors such as diet and lifestyle, and social-environmental factors including living conditions and antibiotic use. Recognizing these interconnections enables development of integrative health strategies that optimize microbiome health as part of comprehensive approaches to disease prevention and health promotion.
Frequently Asked Questions
What exactly is the microbiome?
The microbiome refers to the complete community of microorganisms (bacteria, viruses, fungi, and other microbes) living in and on the human body, along with their genetic material. These microbial communities are essential for human health, contributing to digestion, immune function, and metabolic processes.
How did the Human Microbiome Project change microbiome research?
The Human Microbiome Project revolutionized research by introducing culture-independent methods that allowed scientists to identify microorganisms without requiring laboratory cultivation. This enabled comprehensive mapping of human microbial communities and established baseline data for understanding healthy microbiome composition.
Are all probiotics equally effective?
No, probiotic efficacy varies significantly depending on the specific bacterial strain, dosage, duration of use, and the individual’s existing microbiome and health status. Different strains have different mechanisms of action and benefits, so products should not be assumed to be interchangeable.
What is fecal microbiota transplantation and when is it used?
Fecal microbiota transplantation (FMT) involves transferring stool samples from healthy donors to recipients to restore microbial communities. It is most established for treating recurrent Clostridioides difficile infections, though researchers are exploring its application for other conditions.
How do probiotics affect immune function?
Probiotics interact with intestinal cells and immune tissues, promoting beneficial immune responses. They can enhance innate immunity, promote regulatory T cell development to reduce inflammation, and strengthen intestinal barrier function, though specific effects depend on the probiotic strain.
What are omics technologies in microbiome research?
Omics technologies (genomics, metagenomics, transcriptomics, proteomics, metabolomics) are integrated analytical approaches that examine entire biological systems simultaneously, providing comprehensive insights into microbiome composition, function, and interactions with host physiology.
References
- The Microbiome and the Human Microbiome Project — National Center for Complementary and Integrative Health (NCCIH), National Institutes of Health. 2024. https://www.nccih.nih.gov/training/videolectures/13/4
- NCCIH Strategic Plan FY 2021–2025 — National Center for Complementary and Integrative Health (NCCIH), National Institutes of Health. 2021. https://www.nccih.nih.gov/about/nccih-strategic-plan-2021-2025
- Online Continuing Education Series — National Center for Complementary and Integrative Health (NCCIH), National Institutes of Health. 2024. https://www.nccih.nih.gov/training/videolectures
- Probiotics, Prebiotics, and Synbiotics: Homeostatic Control of the Gut Ecosystem — International Scientific Association for Probiotics and Prebiotics. 2017. https://www.isappscience.org
- Video Library for Researchers — National Center for Complementary and Integrative Health (NCCIH), National Institutes of Health. 2024. https://www.nccih.nih.gov/grants/videos
Read full bio of medha deb